 SageDDA: Extremely fast with minimal memory usage for Complex Data Searches
SageDDA: Extremely fast with minimal memory usage for Complex Data Searches
(Available through the Chaparral platform or as a standalone binary)
At the heart of modern proteomics research is the need for speed and efficiency when analyzing large datasets. SageDDA delivers on both fronts, offering unparalleled performance and reliability for complex data searches. Whether you're dealing with HLA/immunopeptidomics, metaproteomics, microbiome studies, or any large-scale proteomics datasets, SageDDA is specifically designed to meet the challenges of high-throughput proteomics data generated by modern mass spectrometry instruments. SageDDA is built on top of the foundation provided by the open-source version of Sage - keeping Sage's reliability and performance while enabling us to add critical features missing in the open-source version, like automatic parameter optimization and recalibration.
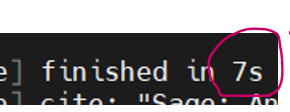
A mere 7 seconds to search and LFQ - combined!!
We benchmarked SageDDA with ~1G Thermo raw file with UniPro human database. If you
Key Features
- Complete Rewrite of Sage (Open Source): Fully redesigned from the ground up to achieve superior speed, memory efficiency, and consistent, stable results.
- Optimized Semi-Supervised Learning Framework: The algorithm behind the widely-used mokapot framework has been reimplemented in Rust, offering significantly improved performance and scalability.
- Robust Global FDR Control: Engineered to support accurate and scalable False Discovery Rate (FDR) estimation across large datasets, enabling confident peptide/protein identification in high-throughput studies.
Proven Efficiency of SageDDA
SageDDA excels in processing high-throughput data rapidly, even when tackling some of the most demanding bioinformatics tasks, such as HLA database searching. The benchmarks presented here, generated using public data from the Carr lab (MassIVE MSV000084172), demonstrate why SageDDA is an essential tool for any researcher:
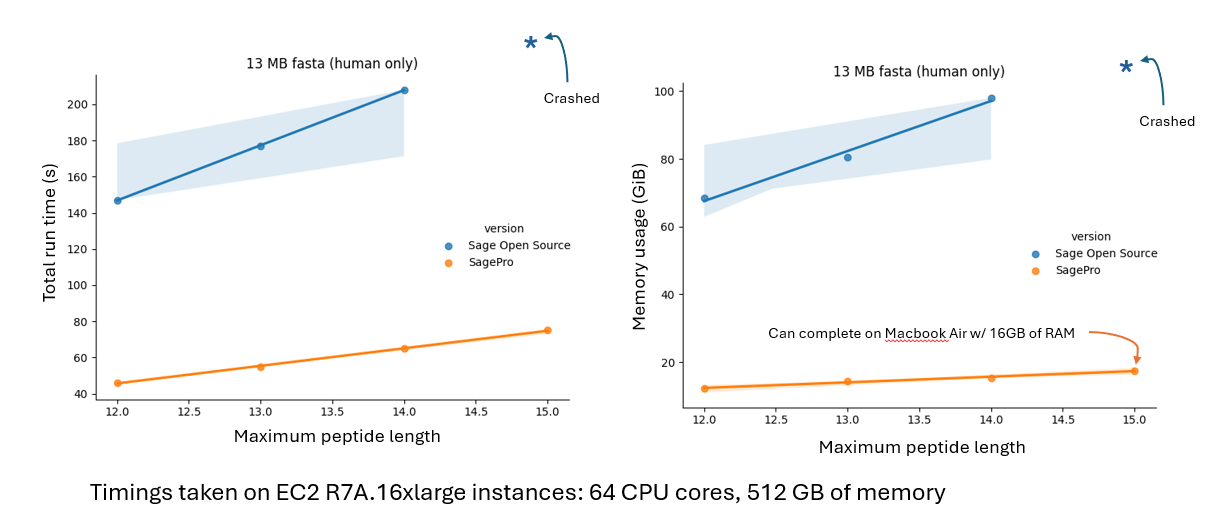
Figure 1: SageDDA Enables Rapid HLA Database Searching
SageDDA was benchmarked against the open-source version of Sage using a UniProt protein database file (human only) for HLA database searches.
- Total Run Time: SageDDA consistently outperforms, maintaining low run times even with the longest peptides. In contrast, Sage Open Source struggles, with increasing run times and system crashes at higher peptide lengths.
- Memory Usage: While Sage Open Source's memory usage increases sharply, leading to crashes, SageDDA remains stable with much lower memory demands. Remarkably, SageDDA completes tasks efficiently on a MacBook Air with 16GB of RAM, while the benchmark setup used much more powerful EC2 R7A.16xlarge instances.
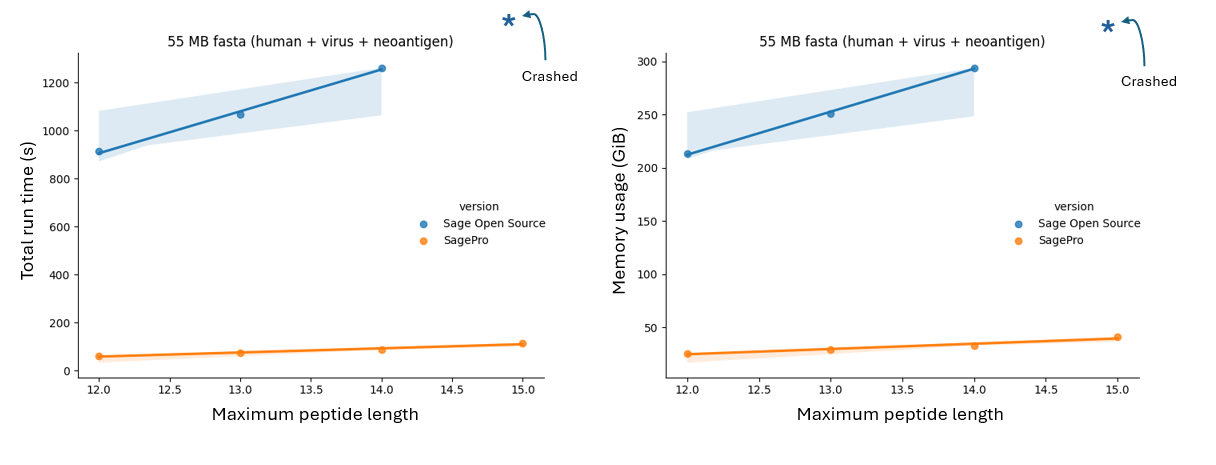
Figure 2: SageDDA Scales Linearly and Predictably
When tested on a larger 55 MB FASTA file (human + virus + neoantigen), SageDDA's superior scalability was evident:
- Total Run Time: SageDDA continues to outperform the open source version of Sage, maintaining low run times even as database sizes grow significantly. Sage Open Source, however, shows substantial run-time increases and crashes at longer peptide lengths.
- Memory Usage: SageDDA's memory usage remains stable even as the database grows, handling extensive datasets without issue. Sage Open Source, on the other hand, crashes under similar conditions.
Why SageDDA is the Ideal Tool for Complex Data Searches
SageDDA's efficiency is particularly critical for HLA/immunopeptidomics data, which require no-enzyme searches, greatly expanding the search space. Its robustness ensures that even the most extensive datasets are processed quickly and reliably. While the examples focus on HLA data, SageDDA's superior run-time and memory performance extend across various datasets, making it applicable to any research scenario requiring high-efficiency data processing.
By choosing SageDDA, you are opting for a tool that not only speeds up your workflows but also ensures stable performance, allowing you to focus on generating insights and breakthroughs from your data. Feel free to reach out to us for inquiries about our software or potential collaborations.